- 20 Oct
- 2025
DETEKSI DINI ABNORMALITAS SEL DARAH PUTIH MENGGUNAKAN ATTENTIVE CNN DENGAN CLASS WEIGHT DAN FOCAL LOSS
ABSTRAK
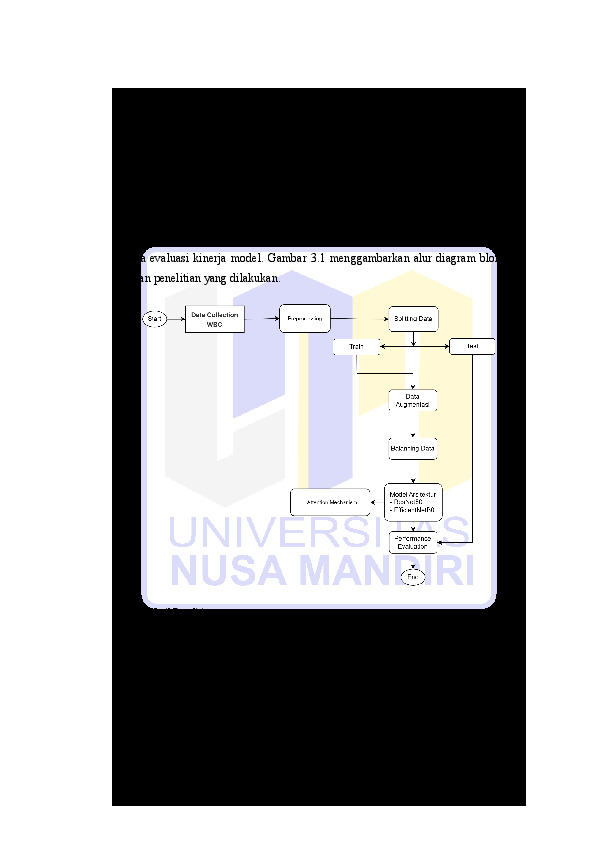
Deteksi dini sel darah putih (WBC) abnormal, seperti leukemia, penting untuk mendukung diagnosis hematology yang akurat. Namun, identifikasi manual melalui citra mikroskopis memakan waktu relative lama dan terkendala oleh tingginya kemiripan morfologi antar sel, sehingga menyulitkan identifikasi secara konsisten. Penelitian ini mengembangkan model klasifikasi WBC berbasis Deep Learning dengan penanganan ketidakseimbangan data dan penerapan Attention Mechanism. Model menggunakan backbone ResNet50 dan EfficientNetB0 dengan teknik balancing Class Weighting dan Focal Loss serta Block Attention CBAM untuk menyoroti fitur morfologi sel. Data diambil dari Munich University Hospital dengan tahap segmentasi ROI, augmentasi dan evaluasi akurasi, F1-Score serta confusion matrix. Hasil menunjukkan akurasi model ResNet50 mencapai 95.86% dengan Macro F1-Score 75.15%, serta peningkatan deteksi pada kelas abnormal. Pendekatan ini mendukung pengembangan sistem klasifikasi otomatis berbasis citra digital untuk diagnosis hematology yang lebih cepat dan andal.
ABSTRACT
Early detection of abnormal white blood cells (WBCs), such as in leukemia, is crucial for supporting accurate hematological diagnosis. However, manual identification through microscopic images is relatively time-consuming and constrained by the high morphological similarity between cell, making it difficult to identify consistently. This study develops a deep learning-based WBC classification model that addresses data imbalance and incorporates an attention mechanism. The model utilizes ResNet50 and EfficientNetB0 backbones, employing Class Weighting and Focal Loss for balancing, as well as the CBAM attention block to emphasize relevant cellular morphological features. The dataset was obtained from Munich University Hospital and underwent ROI segmentation, data augmentation, and evaluation using accuracy, F1-score, and confusion matrix. Results show that the ResNet50 model achieved an accuracy of 95,86% with a macro F1-score 75.15%, demonstrating improved detection performance for abnormal classes. This approach contributes to the development of reliable and efficient image-based automatic classification systems for hematological diagnostics.
Unduhan
REFERENSI
DAFTAR REFERENSI
[1] F. Ernawan, K. Handayani, M. Fakhreldin, and Y. Abbker, “Light Gradient Boosting with Hyper Parameter Tuning Optimization for COVID-19 Prediction,” Int. J. Adv. Comput. Sci. Appl., vol. 13, no. 8, pp. 514–523, 2022, doi: 10.14569/IJACSA.2022.0130859.
[2] O. J. Kweon, Y. K. Lim, M. K. Lee, and H. R. Kim, “Red and white blood cell morphology characterization and hands-on time analysis by the digital cell imaging analyzer DI-60,” PLoS One, vol. 17, no. 4 April, pp. 1–13, 2022, doi: 10.1371/journal.pone.0267638.
[3] M. O. Nakashima, S. N. Coulter, B. J. Blond, R. W. Brown, and J. A. Vos, “The Impact of Pathologist Review on Peripheral Blood Smears: A College of American Pathologists Q-Probes Study of 22 Laboratories,” Arch. Pathol. Lab. Med., vol. 149, no. 7, pp. 618–626, 2025, doi: 10.5858/arpa.2024-0117-cp.
[4] Y. M. Kobara, I. J. Akpan, A. D. Nam, F. H. AlMukthar, and M. Peter, “Artificial Intelligence and Data Science Methods for Automatic Detection of White Blood Cells in Images,” J. Imaging Informatics Med., 2025, doi: 10.1007/s10278-025-01538-y.
[5] M. Dorn et al., “Comparison of machine learning techniques to handle imbalanced COVID-19 CBC datasets,” PeerJ Comput. Sci., vol. 7, pp. 1–34, 2021, doi: 10.7717/peerj-cs.670.
[6] B. Z. Katz, D. Benisty, Y. Sayegh, I. Lamm, and I. Avivi, “Remote Digital Microscopy Improves Hematology Laboratory Workflow by Reducing Peripheral Blood Smear Analysis Turnaround Time,” Appl. Clin. Inform., vol. 13, no. 5, pp. 1108–1115, 2022, doi: 10.1055/a-1957-6219.
[7] S. Dasariraju, M. Huo, and S. McCalla, “Detection and classification of immature leukocytes for diagnosis of acute myeloid leukemia using random forest algorithm,” Bioengineering, vol. 7, no. 4, pp. 1–12, 2020, doi: 10.3390/bioengineering7040120.
[8] T. A. M. Elhassan, M. S. M. Rahim, T. T. Swee, S. Z. M. Hashim, and M. Aljurf, “Feature Extraction of White Blood Cells Using CMYK-Moment Localization and Deep Learning in Acute Myeloid Leukemia Blood Smear Microscopic Images,” IEEE Access, vol. 10, pp. 16577–16591, 2022, doi: 10.1109/ACCESS.2022.3149637.
[9] Siska Rahmadani, Agus Subekti, and M. Haris, “Improving Classification Performance on Imbalanced Medical Data using Generative Adversarial Network,” J. Ilmu Komput. dan Inf., vol. 17, no. 1, pp. 9–17, 2024, doi: 10.21609/jiki.v17i1.1177.
[10] C. Singh, “Medical Imaging using Deep Learning Models,” Eur. J. Eng. Technol. Res., vol. 6, no. 5, pp. 156–167, 2021, doi: 10.24018/ejers.2021.6.5.2491.
[11] A. Batool and Y. C. Byun, “Lightweight EfficientNetB3 Model Based on Depthwise Separable Convolu
tions for Enhancing Classification of Leukemia White Blood Cell Images,” IEEE Access, vol. 11, no. April, pp. 37203–37215, 2023, doi: 10.1109/ACCESS.2023.3266511.
[12] I. H. Sarker, “Machine Learning: Algorithms, Real-World Applications and Research Directions,” SN Comput. Sci., vol. 2, no. 3, pp. 1–21, 2021, doi: 10.1007/s42979-021-00592-x.
[13] Ž. Vujović, “Classification Model Evaluation Metrics,” Int. J. Adv. Comput. Sci. Appl., vol. 12, no. 6, pp. 599–606, 2021, doi: 10.14569/IJACSA.2021.0120670.
[14] H. F. Pardede et al., “Plant diseases detection with low resolution data using nested skip connections,” J. Big Data, vol. 7, no. 1, 2020, doi: 10.1186/s40537-020-00332-7.
[15] A. Aksoy, “An Innovative Hybrid Model for Automatic Detection of White Blood Cells in Clinical Laboratories,” Diagnostics, vol. 14, no. 18, 2024, doi: 10.3390/diagnostics14182093.
[16] B. Ferdinandy et al., “Challenges of machine learning model validation using correlated behaviour data: Evaluation of cross-validation strategies and accuracy measures,” PLoS One, vol. 15, no. 7, pp. 1–14, 2020, doi: 10.1371/journal.pone.0236092.
[17] T. Hasanin, T. M. Khoshgoftaar, J. L. Leevy, and R. A. Bauder, “Severely imbalanced Big Data challenges: investigating data sampling approaches,” J. Big Data, vol. 6, no. 1, 2019, doi: 10.1186/s40537-019-0274-4.
[18] B. BAKIRARAR and A. H. ELHAN, “Class Weighting Technique to Deal with Imbalanced Class Problem in Machine Learning: Methodological Research,” Turkiye Klin. J. Biostat., vol. 15, no. 1, pp. 19–29, 2023, doi: 10.5336/biostatic.2022-93961.
[19] M. Z. Abedin, C. Guotai, P. Hajek, and T. Zhang, “Combining weighted SMOTE with ensemble learning for the class-imbalanced prediction of small business credit risk,” Complex Intell. Syst., vol. 9, no. 4, pp. 3559–3579, 2023, doi: 10.1007/s40747-021-00614-4.
[20] A. Muhammad, Q. Jin, O. Elwasila, and Y. Gulzar, “Hybrid Deep Learning Architecture with Adaptive Feature Fusion for Multi-Stage Alzheimer’s Disease Classification,” Brain Sci., vol. 15, no. 6, pp. 1–23, 2025, doi: 10.3390/brainsci15060612.
[21] O. Katar and I. F. Kilincer, “Automatic Classification of White Blood Cells Using Pre-Trained Deep Models,” Sak. Univ. J. Comput. Inf. Sci., vol. 5, no. 3, pp. 462–476, 2022, doi: 10.35377/saucis...1196934.
[22] R. N. Auer and C. J. Sommer, “Histopathology of Brain Tissue Response to Stroke and Injury,” Stroke Pathophysiol. Diagnosis, Manag., no. November, pp. 0–8, 2021, doi: 10.1016/B978-0-323-69424-7.00004-1.
[23] A. Demir, E. Massaad, and B. Kiziltan, “Topology-Aware Focal Loss for 3D Image Segmentation,” IEEE Comp
- Soc. Conf. Comput. Vis. Pattern Recognit. Work., vol. 2023-June, pp. 580–589, 2023, doi: 10.1109/CVPRW59228.2023.00065.
[24] B. Li et al., “Equalized Focal Loss for Dense Long-Tailed Object Detection,” Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., vol. 2022-June, pp. 6980–6989, 2022, doi: 10.1109/CVPR52688.2022.00686.
[25] M. Yeung, E. Sala, C. B. Schönlieb, and L. Rundo, “Unified Focal loss: Generalising Dice and cross entropy-based losses to handle class imbalanced medical image segmentation,” Comput. Med. Imaging Graph., vol. 95, no. November 2021, 2022, doi: 10.1016/j.compmedimag.2021.102026.
[26] W. Wu and Y. Pan, “Adaptive Modular Convolutional Neural Network for Image Recognition,” Sensors, vol. 22, no. 15, 2022, doi: 10.3390/s22155488.
[27] Y. Wang, X. Chen, J. Li, and Z. Lu, “Convolutional Block Attention Module–Multimodal Feature-Fusion Action Recognition: Enabling Miner Unsafe Action Recognition,” Sensors, vol. 24, no. 14, 2024, doi: 10.3390/s24144557.
[28] M. Zakariah and A. Alnuaim, “Recognizing human activities with the use of Convolutional Block Attention Module,” Egypt. Informatics J., vol. 27, no. September, p. 100536, 2024, doi: 10.1016/j.eij.2024.100536.
[29] S. Wu and H. Yamauchi, “A Lightweight Binarized Convolutional Block Attention Module: B-CBAM,” J. Adv. Inf. Technol., vol. 16, no. 5, pp. 751–759, 2025, doi: 10.12720/jait.16.5.751-759.
[30] D. Maza, J. O. Ojo, and G. O. Akinlade, “A predictive machine learning framework for diabetes,” Turkish J. Eng., vol. 8, no. 3, pp. 583–592, 2024, doi: 10.31127/tuje.1434305.
[31] J. S. Aguilar-Ruiz and M. Michalak, “Classification performance assessment for imbalanced multiclass data,” Sci. Rep., vol. 14, no. 1, pp. 1–10, 2024, doi: 10.1038/s41598-024-61365-z.
[32] S. Uddin, I. Haque, H. Lu, M. A. Moni, and E. Gide, “Comparative performance analysis of K-nearest neighbour (KNN) algorithm and its different variants for disease prediction,” Sci. Rep., vol. 12, no. 1, pp. 1–11, 2022, doi: 10.1038/s41598-022-10358-x.
[33] I. G. Ivanov, Y. Kumchev, and V. J. Hooper, “An Optimization Precise Model of Stroke Data to Improve Stroke Prediction,” Algorithms, vol. 16, no. 9, pp. 1–16, 2023, doi: 10.3390/a16090417.
[34] J. Zhang, L. Chen, and F. Abid, “Prediction of Breast Cancer from Imbalance Respect Using Cluster-Based Undersampling Method,” J. Healthc. Eng., vol. 2019, 2019, doi: 10.1155/2019/7294582.
[35] H. Nizam-Ozogur and Z. Orman, “A heuristic-based hybrid sampling method using a combination of SMOTE and ENN for imbalanced health data,” Expert Syst., vol. 41, no. 8, pp. 1–22, 2024, doi: 10.1111/exsy.13596.
[36] D. Chicco, V. Starovoitov, and G. Jurman, “The Benefits of the Matthews Correlation Coefficient (MCC) over th
e Diagnostic Odds Ratio (DOR) in Binary Classification Assessment,” IEEE Access, vol. 9, no. Mcc, pp. 47112–47124, 2021, doi: 10.1109/ACCESS.2021.3068614.
[37] D. Chicco, M. J. Warrens, and G. Jurman, “The Matthews Correlation Coefficient (MCC) is More Informative Than Cohen’s Kappa and Brier Score in Binary Classification Assessment,” IEEE Access, vol. 9, pp. 78368–78381, 2021, doi: 10.1109/ACCESS.2021.3084050.
[38] H. Song and Z. Wang, “Automatic Classification of White Blood Cells Using a Semi-Supervised Convolutional Neural Network,” IEEE Access, vol. 12, no. February, pp. 44972–44983, 2024, doi: 10.1109/ACCESS.2024.3380896.
[39] J. C. Kiran et al., “White Blood Cells Classification using CNN,” EAI Endorsed Trans. Pervasive Heal. Technol., vol. 9, pp. 1–8, 2023, doi: 10.4108/eetpht.9.4852.
[40] M. S. Abou El-Seoud, M. H. Siala, and G. McKee, “Detection and Classification of White Blood Cells Through Deep Learning Techniques,” Int. J. online Biomed. Eng., vol. 16, no. 15, pp. 94–105, 2020, doi: 10.3991/ijoe.v16i15.15481.
[41] G. Asha, A. Deepthi, B. Sowmya, A. K. Bai, and P. A. Reddy, “Classification of White Blood Cell Images Using Probabilistic Neural Networks,” vol. 3, no. 10, pp. 167–172, 2020.
[42] N. S. Fatonah, H. Tjandrasa, and C. Fatichah, “Identification of acute lymphoblastic leukemia subtypes in touching cells based on enhanced edge detection,” Int. J. Intell. Eng. Syst., vol. 13, no. 4, pp. 204–215, 2020, doi: 10.22266/IJIES2020.0831.18.
[43] J. H. Chien, S. Chan, S. Cheng, and Y. C. Ouyang, “Identification and detection of immature white blood cells through deep learning,” LifeTech 2021 - 2021 IEEE 3rd Glob. Conf. Life Sci. Technol., no. LifeTech, pp. 3–5, 2021, doi: 10.1109/LifeTech52111.2021.9391955.
[44] T. R. Tseng and H. M. Huang, “Classification of peripheral blood neutrophils using deep learning,” Cytom. Part A, vol. 103, no. 4, pp. 295–303, 2023, doi: 10.1002/cyto.a.24698.
[45] B. Hemalatha, B. Karthik, C. V. Krishna Reddy, and A. Latha, “Deep learning approach for segmentation and classification of blood cells using enhanced CNN,” Meas. Sensors, vol. 24, no. September, p. 100582, 2022, doi: 10.1016/j.measen.2022.100582.
[46] D. M. Wonohadidjojo, “Perbandingan Convolutional Neural Network pada Transfer Learning Method untuk Mengklasifikasikan Sel Darah Putih,” Ultim. J. Tek. Inform., vol. 13, no. 1, pp. 51–57, 2021, doi: 10.31937/ti.v13i1.2040.
[47] Mindray, “Atlas of typical blood cells,” Shenzhen Mindray Bio-Medical Electronics Co.,Ltd. All rights reserved. Accessed: Jul. 17, 2025. [Online]. Available: https://www.mindray.com/content/dam/xpace/en/documents/service-tips/hema/typical-blood-cell-atlas-en.pdf
[48] M. L. Chase et al., “Consensus recommendations on peripheral blood smear review: defining curricular standards and fellow competency,” Blood Adv., vol. 7, no. 13, pp. 3244–3252, 2023, doi: 10.1182/bloodadvances.2023009843.
[49] C. Trial et al., “Evaluation of MC-80 automatic blood cell morphology analyzer in identifying the morphology of blood cells in patients with hematological diseases and normal samples,” vol. 29, no. November 2024, 2025.
[50] Y. Zhao, Y. Diao, J. Zheng, X. Li, and H. Luan, “Performance evaluation of the digital morphology analyser Sysmex DI-60 for white blood cell differentials in abnormal samples,” Sci. Rep., vol. 14, no. 1, pp. 1–8, 2024, doi: 10.1038/s41598-024-65427-0.
[51] R. Shasho, “Diagnostic approach to eosinophilia in children,” World Fam. Med. J. /Middle East J. Fam. Med., vol. 19, no. 12, 2021, doi: 10.5742/mewfm.2021.94190.
[52] K. Gu, J. Gao, L. He, Z. Lu, and Y. Zhang, “The role of blood cell morphology in understanding and diagnosing severe fever with thrombocytopenia syndrome (SFTS): Insights from a case report,” Med. (United States), vol. 103, no. 45, p. e40502, 2024, doi: 10.1097/MD.0000000000040502.
[53] N. Merlina, E. Noersasongko, P. N. Andono, M. Arief Soeleman, and D. Riana, “Optimization of the Preprocessing Method for Edge Detection on Overlapping Cells at PAP Smear Images,” Int. J. Informatics Vis., vol. 7, no. 2, pp. 471–476, 2023, doi: 10.30630/joiv.7.2.1329.
[54] D. C. Lepcha, A. Dogra, B. Goyal, V. Goyal, V. Kukreja, and D. P. Bavirisetti, “A constructive non-local means algorithm for low-dose computed tomography denoising with morphological residual processing,” PLoS One, vol. 18, no. 9 September, pp. 1–22, 2023, doi: 10.1371/journal.pone.0291911.
[55] M. Muthumanjula and R. Bhoopalan, “Detection of White Blood Cell Cancer using Deep Learning using Cmyk-Moment Localisation for Information Retrieval,” J. ISMAC, vol. 4, no. 1, pp. 54–72, 2022, doi: 10.36548/jismac.2022.1.006.
[56] R. U. Khan, S. Almakdi, M. Alshehri, A. U. Haq, A. Ullah, and R. Kumar, “An intelligent neural network model to detect red blood cells for various blood structure classification in microscopic medical images,” Heliyon, vol. 10, no. 4, p. e26149, 2024, doi: 10.1016/j.heliyon.2024.e26149.
[57] Q. H. Nguyen et al., “Influence of data splitting on performance of machine learning models in prediction of shear strength of soil,” Math. Probl. Eng., vol. 2021, 2021, doi: 10.1155/2021/4832864.
[58] Z. M. Kouzehkanan et al., “A large dataset of white blood cells containing cell locations and types, along with segmented nuclei and cytoplasm,” Sci. Rep., vol. 12, no. 1, pp. 1–14, 2022, doi: 10.1038/s41598-021-04426-x.
[59] R. Ghorbani, R. Ghousi, A. Makui, and A. Atashi, “A New Hybrid Predictive Model to Predict the Early Mortality Risk in Intensive Care Units on a Highly Imbalanced Dataset,” IEEE Access, vol. 8, pp. 141066–141079, 2020, doi: 10.1109/ACCESS.2020.3013320.
[60] P. Rana, A. Sowmya, E. Meijering, and Y. Song, “Data augmentation with improved regularisation and sampling for imbalanced blood cell image classification,” Sci. Rep., vol. 12, no. 1, pp. 1–13, 2022, doi: 10.1038/s41598-022-22882-x.
[61] X. Gao, R. K. Saha, M. R. Prasad, and A. Roychoudhury, “Fuzz testing based data augmentation to improve robustness of deep neural networks,” Proc. - Int. Conf. Softw. Eng., pp. 1147–1158, 2020, doi: 10.1145/3377811.3380415.
[62] I. Kandel, M. Castelli, and L. Manzoni, “Brightness as an Augmentation Technique for Image Classification,” Emerg. Sci. J., vol. 6, no. 4, pp. 881–892, 2022, doi: 10.28991/ESJ-2022-06-04-015.
[63] L. Nanni, M. Paci, S. Brahnam, and A. Lumini, “Feature transforms for image data augmentation,” Neural Comput. Appl., vol. 34, no. 24, pp. 22345–22356, 2022, doi: 10.1007/s00521-022-07645-z.
[64] M. Toptaş, B. Toptaş, and D. Hanbay, “Classifying white blood cells using combining different convolutional neural networks,” Multimed. Tools Appl., 2025, doi: 10.1007/s11042-025-20879-y.
[65] L. Arora et al., “Ensemble deep learning and EfficientNet for accurate diagnosis of diabetic retinopathy,” Sci. Rep., vol. 14, no. 1, pp. 1–16, 2024, doi: 10.1038/s41598-024-81132-4.