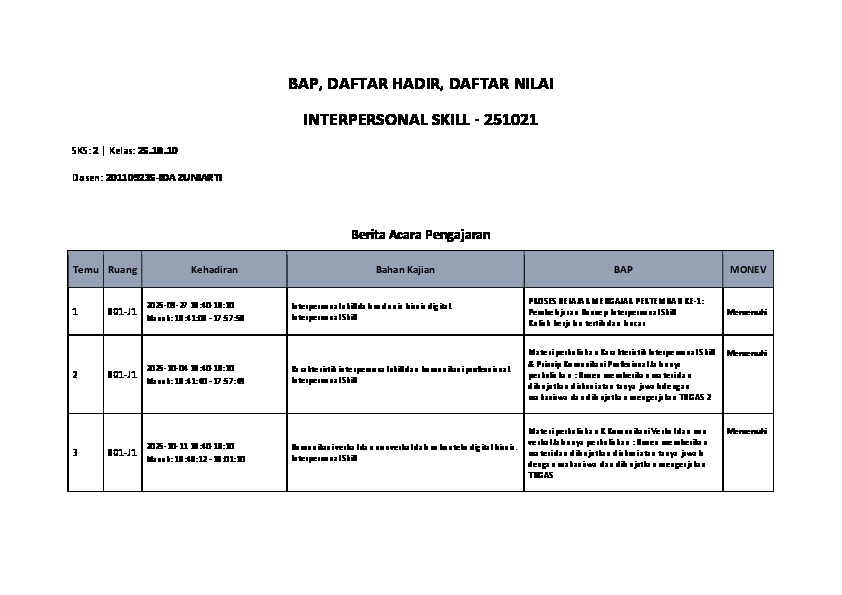
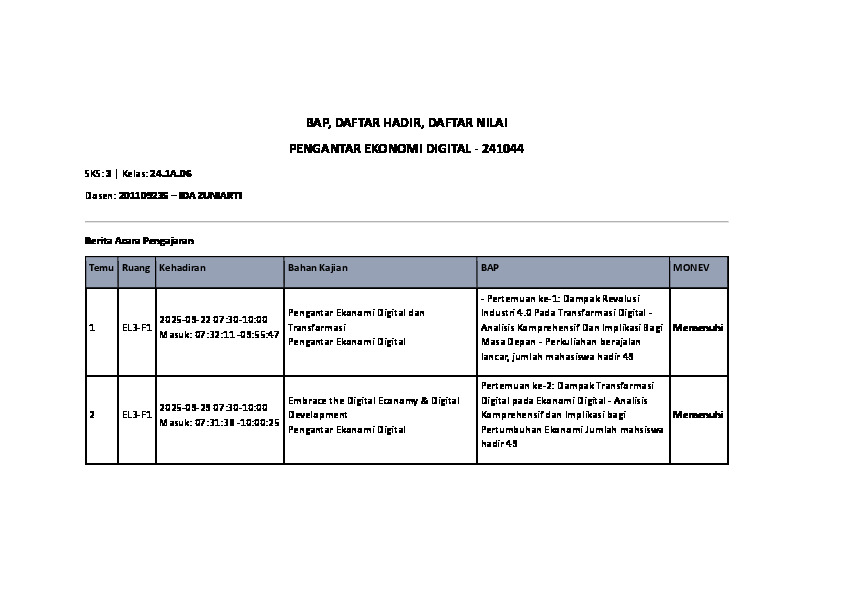
- 08 Mar
- 2025
PREDIKSI KANKER SERVIKS MENGGUNAKAN METODE STACKING ENSEMBLE TABNET, RANDOM FOREST, DAN LIGHT GRADIENT BOOSTING MACHINE
Penelitian ini mengevaluasi akurasi dari beberapa algoritma klasifikasi dalam mendeteksi dini kanker serviks, dengan menjelaskan bahwa Random Fores (RF) menunjukkan kinerja terbaik, dengan mencapai skor akurasi tinggi sebesar 0.978 untuk metode tes skrining deteksi dini kanker serviks Hinselmann dan 0.969 untuk metode tes skrining Biopsy. Praproses dataset dilakukan sebelum penerapan algoritma klasifikasi dilakukan yang meliputi penanganan data yang hilang menggunakan metode Mice Forest Iterative Imputer, serta penanganan ketidakseimbangan kelas dengan teknik Adaptive Synthetic Sampling (ADASYN). Metode seperti Support Vector Machine (SVM) dan K-Nearest Neighbors (KNN) memperlihatkan kinerja yang cukup baik, akan tetapi teknik Ensemble secara significant mampu meningkatkan kemampuan prediksi. Penerapan metode Ensemble Stacking (Tabnet, RF, dan LightGBM) yang mencapai akurasi 1 pada Hinselmann dan Biopsy, ini menunjukkan keefektifitasan dari kombinasi beberapa algoritma. Dengan hasil tersebut, peneliti telah berhasil menunjukkan pentingnya untuk memilih metode klasifikasi yang tepat dalam mendeteksi dini kanker serviks, serta menunjukkan bahwa model Ensemble yang canggih dapat mengarah pada peningkatan prediksi deteksi dini yang lebih baik.
Unduhan
-
File_10 Lampiran.pdf
Terakhir download 08 Apr 2025 08:04File_10 Lampiran
- diunduh 2x | Ukuran 228 KB
-
File_12 Surat Pernyataan.pdf
Terakhir download 19 Feb 2026 09:02File_12 Surat Pernyataan
- diunduh 0x | Ukuran 277 KB
-
File_12 Surat Pernyataan.pdf
Terakhir download 19 Feb 2026 09:02File_12 Surat Pernyataan
- diunduh 0x | Ukuran 283,349
-
File_9 Daftar Referensi.pdf
Terakhir download 19 Feb 2026 09:02File_09 Daftar Referensi
- diunduh 0x | Ukuran 469,559
-
File_13 Tesis Keseluruhan.pdf
Terakhir download 21 Mar 2025 02:03File_13 Tesis Keseluruhan
- diunduh 2x | Ukuran 9,635,577
-
File_1 Cover Halaman Judul Laporan Tesis.pdf
Terakhir download 18 Feb 2026 11:02File_01 Cover
- diunduh 71x | Ukuran 207,850
REFERENSI
- Exner et al., “Value of diffusion-weighted MRI in diagnosis of uterine cervical cancer: a prospective study evaluating the benefits of DWI compared to conventional MR sequences in a 3T environment,” Acta Radiol, vol. 57, no. 7, pp. 869–877, Jul. 2016, doi: 10.1177/0284185115602146.
[2] P. Z. McVeigh, A. M. Syed, M. Milosevic, A. Fyles, and M. A. Haider, “Diffusion-weighted MRI in cervical cancer,” Eur Radiol, vol. 18, no. 5, pp. 1058–1064, May 2008, doi: 10.1007/s00330-007-0843-3.
[3] A. Gadducci, C. Barsotti, S. Cosio, L. Domenici, and A. Riccardo Genazzani, “Smoking habit, immune suppression, oral contraceptive use, and hormone replacement therapy use and cervical carcinogenesis: a review of the literature,” Gynecol Endocrinol, vol. 27, no. 8, pp. 597–604, Aug. 2011, doi: 10.3109/09513590.2011.558953.
[4] P. Luhn et al., “The role of co-factors in the progression from human papillomavirus infection to cervical cancer,” Gynecol Oncol, vol. 128, no. 2, pp. 265–270, Feb. 2013, doi: 10.1016/j.ygyno.2012.11.003.
[5] M. F. A. Razak, M. I. Jaya, F. Ernawan, A. Firdaus, and F. A. Nugroho, “Comparative Analysis of Machine Learning Classifiers for Phishing Detection,” in 2022 6th International Conference on Informatics and Computational Sciences (ICICoS), Sep. 2022, pp. 84–88. doi: 10.1109/ICICoS56336.2022.9930531.
[6] A. H. Elmi, A. Abdullahi, and M. A. Bare, “A comparative analysis of cervical cancer diagnosis using machine learning techniques,” Indonesian Journal of Electrical Engineering and Computer Science, vol. 34, no. 2, Art. no. 2, May 2024, doi: 10.11591/ijeecs.v34.i2.pp1010-1023.
[7] M. M. Ali et al., “Machine learning-based statistical analysis for early stage detection of cervical cancer,” Computers in Biology and Medicine, vol. 139, p. 104985, 2021, doi: https://doi.org/10.1016/j.compbiomed.2021.104985.
[8] C. Janiesch, P. Zschech, and K. Heinrich, “Machine learning and deep learning,” Electronic Markets, vol. 31, no. 3, pp. 685–695, Sep. 2021, doi: 10.1007/s12525-021-00475-2.
[9] J. Ogunleye, “The Concept of Data Mining,” 2021. doi: 10.5772/intechopen.99417.
[10] M. D. Samad, S. Abrar, and N. Diawara, “Missing value estimation using clustering and deep learning within multiple imputation framework,” Knowledge-Based Systems, vol. 249, p. 108968, 2022, doi: https://doi.org/10.1016/j.knosys.2022.108968.
[11] T. Xu, G. Coco, and M. Neale, “A predictive model of recreational water quality based on adaptive synthetic sampling algorithms and machine learning,” Water Research, vol. 177, p. 115788, Apr. 2020, doi: 10.1016/j.watres.2020.115788.
[12] M. Mera-Gaona, D. López, R. Vargas, and U. Neumann, “Framework for the Ensemble of Feature Selection Methods,” Applied Sciences, vol. 11, p. 8122, Sep. 2021, doi: 10.3390/app11178122.
[13] D. Mienye and Y. Sun, “A Survey of Ensemble Learning: Concepts, Algorithms, Applications, and Prospects,” IEEE Access, vol. PP, pp. 1–1, Sep. 2022, doi: 10.1109/ACCESS.2022.3207287.
[14] J.-M. Nguyen et al., “Random forest of perfect trees: concept, performance, applications and perspectives.,” Bioinformatics, vol. 37, no. 15, pp. 2165–2174, Aug. 2021, doi: 10.1093/bioinformatics/btab074.
[15] E. Fitri and D. Riana, “ANALISA PERBANDINGAN MODEL PREDICTION DALAM PREDIKSI HARGA SAHAM MENGGUNAKAN METODE LINEAR REGRESSION, RANDOM FOREST REGRESSION DAN MULTILAYER PERCEPTRON,” METHOMIKA: Jurnal Manajemen Informatika & Komputerisasi Akuntansi, vol. 6, no. 1, Art. no. 1, Apr. 2022, doi: 10.46880/jmika.Vol6No1.pp69-78.
[16] D. Gunawan, D. Riana, D. Ardiansyah, and F. Akbar, “Komparasi Algoritma Support Vector Machine Dan Naïve Bayes Dengan Algotima Genetika Pada Analisis Sentimen Calon Gubernur Jabar 2018-2023,” Jurnal Teknik Komputer, vol. 6, pp. 121–129, Jan. 2020, doi: 10.31294/jtk.v6i1.6866.
[17] E. K. Sahin, “Assessing the predictive capability of ensemble tree methods for landslide susceptibility mapping using XGBoost, gradient boosting machine, and random forest,” SN Appl. Sci., vol. 2, no. 7, p. 1308, Jul. 2020, doi: 10.1007/s42452-020-3060-1.
[18] W. Li, Y. Chen, and Y. Song, “Boosted K-nearest neighbor classifiers based on fuzzy granules,” Knowledge-Based Systems, vol. 195, p. 105606, 2020, doi: https://doi.org/10.1016/j.knosys.2020.105606.
[19] H. Saputra, A. Efendi, E. Rudini, D. Riana, and A. Hewiz, “Hepatitis Prediction Using K-NN, Naive Bayes, Support Vector Machine, Multilayer Perceptron and Random Forest, Gradient Boosting, K-Means,” Journal Medical Informatics Technology, pp. 96–100, Dec. 2023, doi: 10.37034/medinftech.v1i4.21.
[20] N. Nurajijah and D. Riana, “Algoritma Naïve Bayes, Decision Tree, dan SVM untuk Klasifikasi Persetujuan Pembiayaan Nasabah Koperasi Syariah,” Jurnal Teknologi dan Sistem Komputer, vol. 7, p. 77, Apr. 2019, doi: 10.14710/jtsiskom.7.2.2019.77-82.
[21] E. Christodoulou, J. Ma, G. Collins, E. Steyerberg, J. Verbakel, and B. Van Calster, “A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models,” Journal of Clinical Epidemiology, vol. 110, Feb. 2019, doi: 10.1016/j.jclinepi.2019.02.004.
[22] S. Arik and T. Pfister, “TabNet: Attentive Interpretable Tabular Learning,” Proceedings of the AAAI Conference on Artificial Intelligence, vol. 35, pp. 6679–6687, May 2021, doi: 10.1609/aaai.v35i8.16826.
[23] F. Ernawan, K. Handayani, M. Fakhreldin, and Y. Abbker, “Light Gradient Boosting with Hyper Parameter Tuning Optimization for COVID-19 Prediction,” International Journal of Advanced Computer Science and Applications, vol. 13, no. 8, 2022, doi: 10.14569/IJACSA.2022.0130859.
[24] J. Zhang, D. Mucs, U. Norinder, and F. Svensson, “LightGBM: An Effective and Scalable Algorithm for Prediction of Chemical Toxicity-Application to the Tox21 and Mutagenicity Data Sets.,” J Chem Inf Model, vol. 59, no. 10, pp. 4150–4158, Oct. 2019, doi: 10.1021/acs.jcim.9b00633.
[25] A. Islam, S. Ripon, and Md. N. Bhuiyan, “Cervical Cancer Risk Factors: Classification and Mining Associations,” APTIKOM Journal on Computer Science and Information Technologies, vol. 4, pp. 8–18, Jan. 2020, doi: 10.34306/csit.v4i1.85.
[26] W. Yang, X. Gou, T. Xu, X. Yi, and M. Jiang, “Cervical Cancer Risk Prediction Model and Analysis of Risk Factors based on Machine Learning,” in Proceedings of the 2019 11th International Conference on Bioinformatics and Biomedical Technology, in ICBBT’19. New York, NY, USA: Association for Computing Machinery, Mei 2019, pp. 50–54. doi: 10.1145/3340074.3340078.
[27] X. Deng, Y. Luo, and C. Wang, “Analysis of Risk Factors for Cervical Cancer Based on Machine Learning Methods,” in 2018 5th IEEE International Conference on Cloud Computing and Intelligence Systems (CCIS), Nov. 2018, pp. 631–635. doi: 10.1109/CCIS.2018.8691126.
[28] A. AlMohimeed, H. Saleh, S. Ibrahim, R. Saad, and A. Samy, “Cervical Cancer Diagnosis Using Stacked Ensemble Model and Optimized Feature Selection: An Explainable Artificial Intelligence Approach,” Computers, vol. 12, p. 200, Oct. 2023, doi: 10.3390/computers12100200.
[29] N. A. Mudawi and A. Alazeb, “A Model for Predicting Cervical Cancer Using Machine Learning Algorithms,” Sensors (Basel, Switzerland), vol. 22, no. 11, p. 4132, May 2022, doi: 10.3390/s22114132.
[30] Md. A. Rahman and A. Ghosh, “Risk factor analysis and prediction of Cervical Cancer based on machine learning models,” Dec. 2023, pp. 1–4. doi: 10.1109/ICCIT60459.2023.10441011.
[31] A. Dharma et al., “Deteksi Pola Pasien Kanker Serviks dengan Algoritma Extra Trees dan K-Nearest Neighbor,” Jurnal Ilmu Komputer dan Sistem Informasi (JIKOMSI), vol. 3, no. 1, pp. 32–36, Oct. 2020, doi: 10.9767/jikomsi.v3i1.80.
[32] J. C. Kelwin Fernandes, “Cervical cancer (Risk Factors).” UCI Machine Learning Repository, 2017. doi: 10.24432/C5Z310.
[33] E. Slade and M. Naylor, “A fair comparison of tree‐based and parametric methods in multiple imputation by chained equations,” Statistics in Medicine, vol. 39, Jan. 2020, doi: 10.1002/sim.8468.
[34] S. Fotouhi, S. Asadi, and M. W. Kattan, “A comprehensive data level analysis for cancer diagnosis on imbalanced data,” Journal of Biomedical Informatics, vol. 90, p. 103089, 2019, doi: https://doi.org/10.1016/j.jbi.2018.12.003.
[35] C. Ntakolia, C. Kokkotis, S. Moustakidis, and D. Tsaopoulos, “Identification of most important features based on a fuzzy ensemble technique: Evaluation on Joint Space Narrowing Progression in Knee Osteoarthritis Patients,” International Journal of Medical Informatics, vol. 156, p. 104614, Oct. 2021, doi: 10.1016/j.ijmedinf.2021.104614.
[36] P. Hillemanns et al., “Prevention of Cervical Cancer: Guideline of the DGGG and the DKG (S3 Level, AWMF Register Number 015/027OL, December 2017) - Part 2 on Triage, Treatment and Follow-up.,” Geburtshilfe Frauenheilkd, vol. 79, no. 2, pp. 160–176, Feb. 2019, doi: 10.1055/a-0828-7722.
[37] H. Fauzi, G. Surya, R. Magdalena, A. B. Harsono, and T. Azhar, “Sistem Deteksi Pra-Kanker Serviks dengan Pengolahan Citra Hasil Inspeksi Visual Asam Asetat,” Techno.Com, vol. 20, pp. 290–299, May 2021, doi: 10.33633/tc.v20i2.4285.
[38] A. L. Swailes, C. E. Hossler, and J. P. Kesterson, “Pathway to the Papanicolaou smear: The development of cervical cytology in twentieth-century America and implications in the present day.,” Gynecol Oncol, vol. 154, no. 1, pp. 3–7, Jul. 2019, doi: 10.1016/j.ygyno.2019.04.004.
[39] N. Merlina, E. Noersasongko, P. N. Andono, M. A. Soeleman, and D. Riana, “Optimization of the Preprocessing Method for Edge Detection on Overlapping Cells at PAP Smear Images,” JOIV : International Journal on Informatics Visualization, vol. 7, no. 2, pp. 471–476, May 2023, doi: 10.30630/joiv.7.2.1329.
[40] P. Hofman, “The challenges of evaluating predictive biomarkers using small biopsy tissue samples and liquid biopsies from non-small cell lung cancer patients.,” J Thorac Dis, vol. 11, no. Suppl 1, pp. S57–S64, Jan. 2019, doi: 10.21037/jtd.2018.11.85.
[41] K. Moorthy, A. N. Jaber, M. A. Ismail, F. Ernawan, M. S. Mohamad, and S. Deris, “Missing-Values Imputation Algorithms for Microarray Gene Expression Data,” in Microarray Bioinformatics, V. Bolón-Canedo and A. Alonso-Betanzos, Eds., New York, NY: Springer, 2019, pp. 255–266. doi: 10.1007/978-1-4939-9442-7_12.
[42] W. Wu and H. Zhou, “Data-Driven Diagnosis of Cervical Cancer With Support Vector Machine-Based Approaches,” IEEE Access, vol. 5, pp. 25189–25195, 2017, doi: 10.1109/ACCESS.2017.2763984.
[43] N. G. Ramadhan, “Comparative Analysis of ADASYN-SVM and SMOTE-SVM Methods on the Detection of Type 2 Diabetes Mellitus,” Scientific Journal of Informatics, vol. 8, no. 2, Art. no. 2, Nov. 2021, doi: 10.15294/sji.v8i2.32484.
[44] S. Chohan, A. Nugroho, A. Aji, and W. Gata, “Analisis Sentimen Pengguna Aplikasi Duolingo Menggunakan Metode Naïve Bayes dan Synthetic Minority Over Sampling Technique,” Paradigma - Jurnal Komputer dan Informatika, vol. 22, pp. 139–144, Sep. 2020, doi: 10.31294/p.v22i2.8251.
[45] I. Ahmed et al., “Eye Tracking-Based Diagnosis and Early Detection of Autism Spectrum Disorder Using Machine Learning and Deep Learning Techniques,” Electronics, vol. 11, p. 530, Feb. 2022, doi: 10.3390/electronics11040530.
[46] P. Singh, G. Pal, and S. Gangwar, “Prediction of Cardiovascular Disease Using Feature Selection Techniques,” International Journal of Computer Theory and Engineering, vol. 14, pp. 97–103, Jan. 2022, doi: 10.7763/IJCTE.2022.V14.1316.
[47] M. Anasanti, K. Hilyati, and A. Novtariany, “The Exploring feature selection techniques on Classification Algorithms for Predicting Type 2 Diabetes at Early Stage,” Jurnal RESTI (Rekayasa Sistem dan Teknologi Informasi), vol. 6, pp. 832–839, Nov. 2022, doi: 10.29207/resti.v6i5.4419.